Hi all,
Last week I wrote a small MATLAB code to compute the result of some theoretical calculations I made.
These calculations are aimed to extract the spectral function A(\mathbf{k},\omega) from the Green’s function (\mathcal{G}) derived using Dyson’s equation and the self-energy (\Sigma).
The purpose of the code Is to compute the self energy (product of know matrices), then the Green’s function and finally extract the spectral density:
with \epsilon_\mathbf{k} single-particle unperturbed energy and \mathcal{G}_0 non-interacting Green’s Function.
When it came to testing the code, I set the interaction potential to be zero, so that \mathcal{G}\rightarrow \mathcal{G}_0. However, the result was something like this:
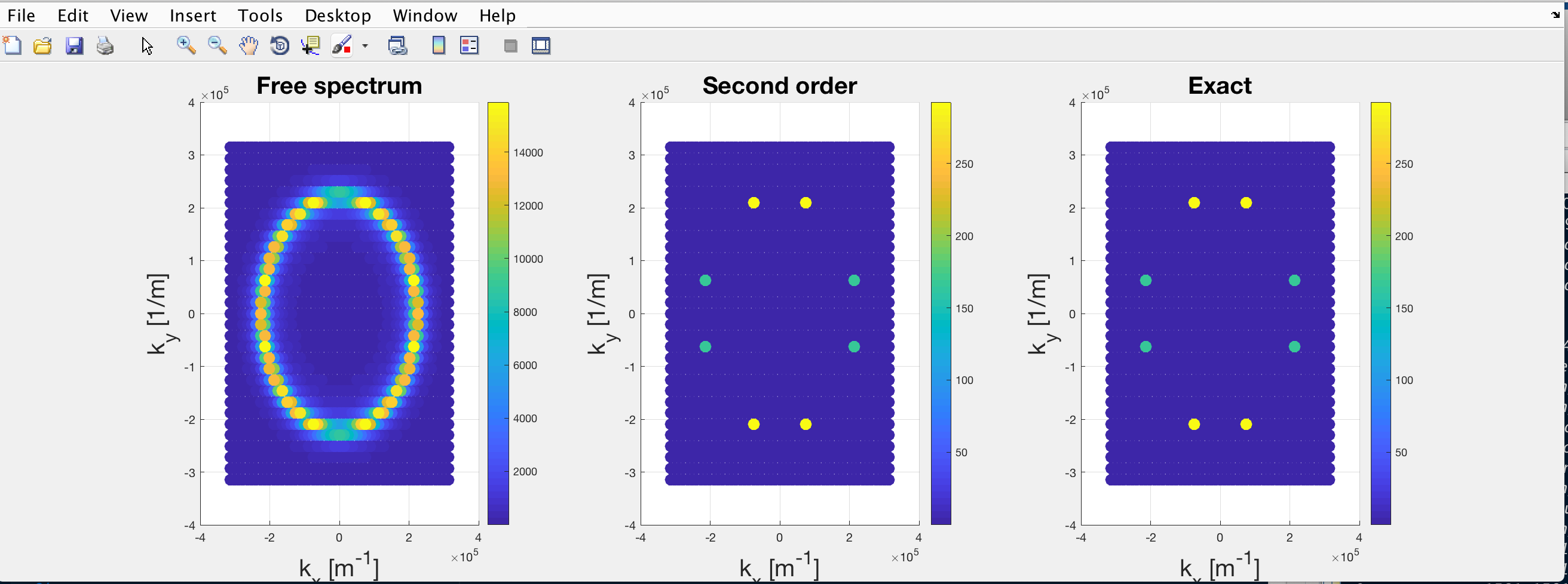
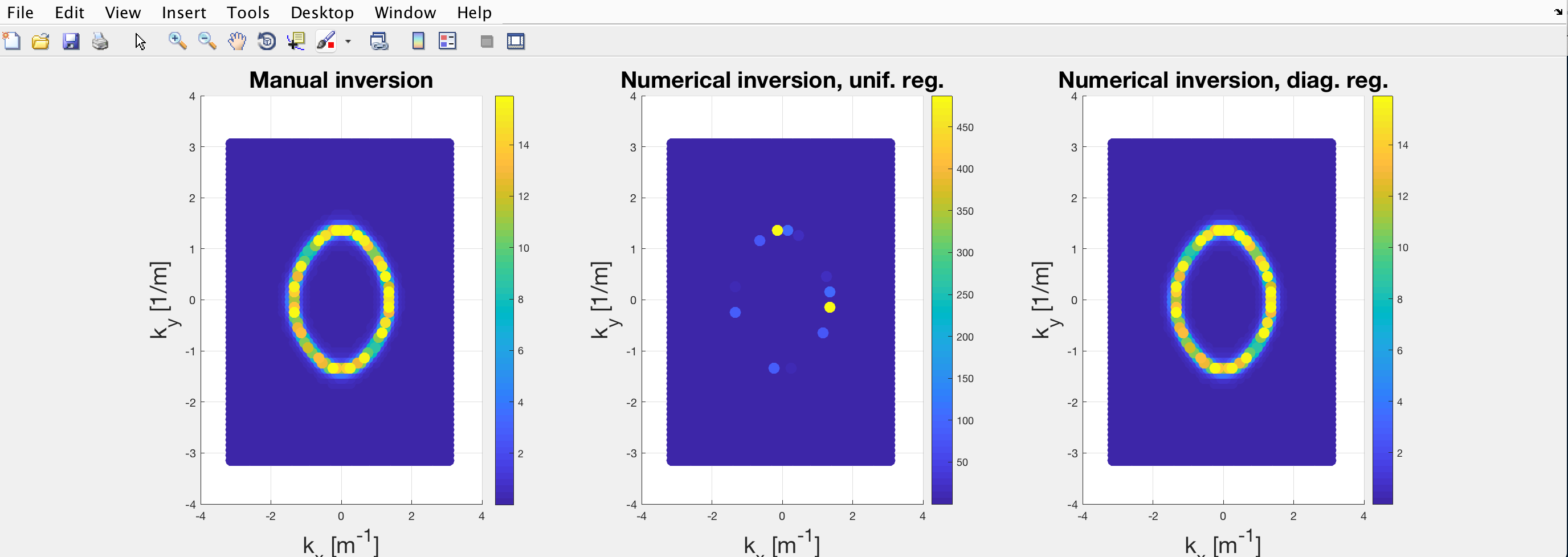
where the colorbar is proportional to A(\mathbf{k}, \omega).
The leftmost plot is derived from the “manual” inversion of \mathcal{G}_0 (replaced the each diagonal element with its inverse) and represents the normal parabolic dispersion of the noninteracting modes. On the other hand, the central and rightmost plots are both derived from the numerical inversion of (\mathcal{G}_{0}^{-1}-\Sigma) which is diagonal as \Sigma=0. Since the interaction is null, the three plots should be identical.
I tracked down this issue to the matrix inversion function required to compute \mathcal{G} .
I understand this being a consequence of the size of the matrix (thousands by thousands of elements) which causes the determinant to easily exceed what can be stored in a double.I tried several different functions pinv(), \ and inv() which gave all the same unsatisfactory result.
Therefore my questions are:
- Could there be some benefit by moving the code to a different language (Python, C)?
- Could it be beneficial to use an external computational cluster to solve the issue?
- Is there a more clever way of addressing the problem instead of inverting a huge matrix?
Apologies for the lengthy question but I felt that it needed some context.
2020-08-21T22:00:00Z